Population genomic analysis unravels the evolutionary processes leading to budding speciation
- 1. National Engineering Laboratory for Resource Development of Endangered Crude Drugs in Northwest China, College of Life Sciences, Shaanxi Normal University, Xi'an 710119, China
2. Key Laboratory of Medicinal Plant Resource and Natural Pharmaceutical Chemistry of Ministry of Education, Shaanxi Normal University, Xi'an 710119, China
†These authors contributed equally to this work.
*Correspondence: Jian‐Qiang Zhang (jqzhang@snnu.edu.cn)
Received date: 2024-09-30
Accepted date: 2025-03-07
Online published: 2025-04-01
Supported by
This work was supported by the National Natural Science Foundation of China (grant nos.: 32070236 and 32370226), the Innovation Capability Support Program of Shaanxi (No. 2023KJXX‐019), and the Fundamental Research Funds for the Central Universities (No. GK202301008 to J.Q. Zhang, and No. LHRCCX23183 to L. Huang).
Abstract
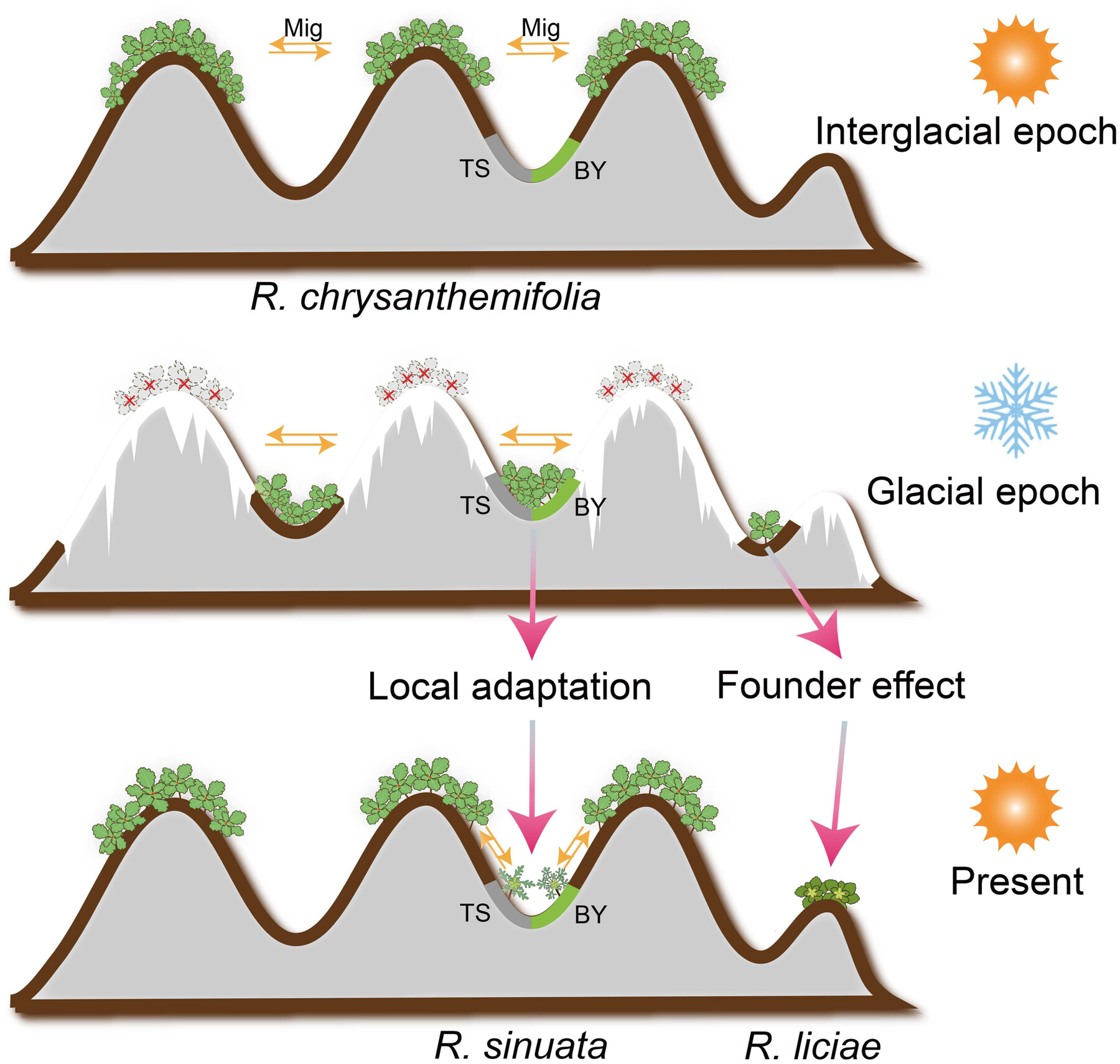
Cite this article
Xiao-Ying Liu, Long Huang, Ya-Peng Yang, Yue-Yi Li, Zi-Wei Ma, Shi-Yu Wang, Lin-Feng Qiu, Qing-Song Liu, Jian-Qiang Zhang . Population genomic analysis unravels the evolutionary processes leading to budding speciation[J]. Journal of Integrative Plant Biology, 0 : 1 . DOI: 10.1111/jipb.13905